The ABC of DNA Computing
For web information about DNA consult the Human Genome Project page at Oak Ridge. Nice images available from The Electron Micrograph Laboratory at the University of Wisconsin. For information about NP-complete problems try Forbes Lewis' page at the University of Kentucky and John Morris' course notes at the University of Western Australia. A useful print reference is DNA Computing by Paun, Rozenberg and Salomaa (Springer, 1998).
1. What is DNA?
DNA (Deoxyribonucleic acid) is the molecular basis of genetics. For our purposes, the following features are important.
Complementarity, the fundamental key to DNA replication, is also the basic mathematical ingredient in DNA computing. --Tony Phillips
SUNY at Stony Brook
*(added November 2015) Actually, the convention is for a string like ATCG to represent 5'-ATCG-3'. Thanks to Gos Micklem for bringing this to our attention.
Welcome to the
Feature Column!
These web essays are designed for those who have already discovered the joys of mathematics as well as for those who may be uncomfortable with mathematics.
Read more . . .
Feature Column at a glance


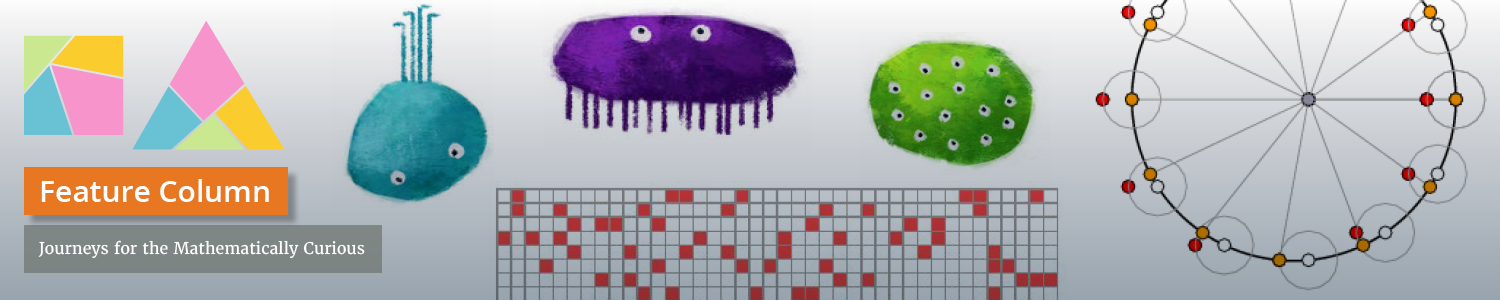
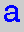 = adenine,
= adenine, 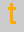 = thymine,
= thymine,  = cytosine,
= cytosine,  = guanine.
= guanine.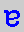 ,
,  ,
,  ,
,  when the strand is being read the opposite way, that is, from the 5'-end to the 3'-end.*
when the strand is being read the opposite way, that is, from the 5'-end to the 3'-end.*